11 KiB
Executable File
marp
| marp |
|---|
| true |
caret & bigstatsR Workshop
Required Libraries
# "bigstatsR" and related packages.
install.packages("bigstatsr")
install.packages("bigreadr")
install.packages("data.table")
# "impute" library.
install.packages("BiocManager")
BiocManager::install("impute")
# "caret" library and sample models.
install.packages("caret")
install.packages("glmnet")
Getting Started
-
All files used in the workshop can be downloaded here:
-
Throughout the workshop, you will need to fill out the missing code in the scripts, marked as
(...).
bigstatsR Package
Provides functions for fast statistical analysis of large-scale data encoded as matrices. The package can handle matrices that are too large to fit in memory thanks to memory-mapping to binary files on disk.
Reading Files with bigstatsR [1]
# Read only the first row of the file.
# This allows us to obtain the number of columns.
first_row <- fread2("Test_Genotype_Data.csv",
nrows = 1)
col_num <- ncol(first_row)
# Read the entire file using big_read.
gen_data <- big_read("Test_Genotype_Data.csv", # Our large data file.
select = 1:col_num, # The number of columns we want to read.
backingfile = "Data", # Path to the backing file.
progress = TRUE, # Show progress.
type = "double") # Data type of the contents.
Reading Files with bigstatsR [2]
- Our file will be read as a Filebacked Big Matrix (FBM).
- After executing the code, we should see the following:
> gen_data
A Filebacked Big Matrix of type 'double' with 1000 rows and 1000 columns.
- Be careful to remove the backing files before trying to re-read the same file.
Working with FBMs [1]
- We can interact with FBMs in a similar manner as we do with normal dataframes.
# Examine the data.
gen_data
gen_data[1:10, 1:10]
str(gen_data)
Working with FBMs [2]
- Functions need to be applied differently.
- Say we wanted to find out the number of missing values in our data.
- The following approach does not work anymore:
# Will not work!
sum(is.na(gen_data))
Applying Functions [1]
- In order for the function to be applied, we need to define it separately and use
big_apply. - Our function must take the following as arguments:
- An FBM object:
X - A vector of indices:
ind
- An FBM object:
# Function for checking the number of missing values in the dataframe.
check_na <- function(X, ind) {
sum(is.na(X[, ind]))
}
big_applywill take care of splitting our data into subsets, applying our function to each subset, and combining the results.
Applying Functions [2]
- Now, we can find the number of missing values in our dataset.
# Check how many missing values we have.
big_apply(gen_data, # FBM to which we want to apply the function.
a.FUN = check_na, # The function that we want to apply.
a.combine = "+", # How the results of each subset should be combined.
ncores = 2) # Number of CPU cores to use.
Applying Functions [3]
- If we really want to, we can manually create subsets and apply functions using a
forloop. - First, we must get the indices for each subset:
# Subset the dataframe into sets of 100 columns.
sub_idx <- split(1:col_num, ceiling(seq_along(1:col_num) / 100))
Applying Functions [4]
- Now, we can extract our subsets and apply our function:
for (i in 1:length(sub_idx)) {
# Display current subset being evaluated.
print(i)
# Get the Genotype Data for the current subset.
gen_subset <- gen_data[, sub_idx[[i]]]
# Impute missing data using KNN clustering.
gen_imputed <- round(
t(impute::impute.knn(t(gen_subset), k = 50, rowmax = 1, colmax = 1)$data), 0)
# "Write back" imputed data to the FBM.
gen_data[, sub_idx[[i]]] <- gen_imputed
}
Applying Functions [5]
- Check that the
imputefunction worked:
# Now, verify there are no missing values anymore.
big_apply(gen_data,
a.FUN = check_na,
a.combine = "+",
ncores = 2)
Saving FBMs
- How do we save our modified data back to a
.csvfile? Easy, usebig_write:
# Write the newly-imputed data to a new file.
big_write(gen_data, # Our FBM object.
"Test_Genotype_Data_Imputed.csv", # Name of the file.
every_nrow = 100, # How many rows do we want to save at once.
progress = interactive()) # Show a fancy progress bar.
Removing Backing Files
big_readwill create backing files that are of no use after we saved our file.- They can be safely removed:
# Remove backing files.
system("rm *.bk")
system("rm *.rds")
caret Package
Short for Classification And REgression Training. It is a set of functions that attempt to streamline the process for creating predictive models. The package started off as a way to provide a uniform interface for modeling functions, as well as a way to standardize common tasks (such parameter tuning and variable importance).
Loading Data
- First, let's load our newly-imputed Genotype Data:
## Data Loading [Predictors]
# Genotype Data
gen_path <- "Test_Genotype_Data_Imputed.csv"
gen_data <- fread(gen_path, sep = ",", header = FALSE)
- Let's load our Phenotype Data as well:
## Data Loading [Traits]
# Phenotype Data
phn_path <- "Test_Phenotype_Data.csv"
phn_data <- fread(phn_path, sep = ",", header = FALSE)
Data Overview
- Our Genotype Data is a large matrix of $0$s and $1$s:
# Preview of Genotype Data.
str(gen_data)
gen_data[1:10, 1:10]
- Our Phenotype Data, which we want to predict, is a single column of
Plant Height (cm)measurements:
# Preview of Phenotype Data.
str(phn_data)
phn_data[1:10, 1]
The createDataPartition Function [1]
- We can easily split our data into training and testing sets as follows:
# We want 80% of our data for training and 20% for testing.
# Get the indices for the rows to be used in the training data.
train_index <- createDataPartition(
phn_data$V1, # Our Phenotype Data.
p = 0.8, # Percentage of data to be used for training.
list = FALSE) # To return a matrix instead of a list.
# Check out which rows will be used for training.
train_index
The createDataPartition Function [2]
- Now that we know which indices will be used for training, we can extract the corresponding data:
# The values used as predictors [x].
x_train <- gen_data[train_index,]
x_test <- gen_data[-train_index,]
# The values to be predicted [y].
y_train <- phn_data[train_index,]
y_test <- phn_data[-train_index,]
Model Training and Tuning [1]
Fits generalized linear and similar models via penalized maximum likelihood. The regularization path is computed for the lasso or elastic net penalty at a grid of values (on the log scale) for the regularization parameter
lambda.
- We can perform this tuning automatically with
caret. Let's first create a custom parameter grid:
# Define a custom tuning grid.
tune_grid <- expand.grid(alpha = seq(0.0001, 1, length = 5), # Values to try for "alpha".
lambda = 5) # Values to try for "lambda".
Model Training and Tuning [2]
- The
trainControlfunction allows us to further customize how our model is made:
# Parameter tuning.
param_tune <- trainControl(method = "repeatedcv", # Method to be used for resampling.
number = 2, # Numebr of folds (subsets) for cross-validation.
repeats = 5, # Number of iterations for cross-validation.
trim = TRUE, # Reduces memory consumption.
search = "grid", # How the tuning grid should be made.
verboseIter = TRUE) # To display additional information.
- Other methods are available, such as a single cross-validation (
cv), a leave-one-out cross-validation (LOOCV), ornone, which will only train a single model. - It is also possible to specify a
randomtuning grid.
Model Training and Tuning [3]
- Now, we can finally train our model:
# Train a model.
glmnet_model <- train(x_train, # Our predictors.
y_train$V1, # What we want to predict.
method = "glmnet", # The model we want to use.
metric = "MAE", # What metric should we use to pick the best model.
tuneGrid = tune_grid, # Our custom tuning grid.
trControl = param_tune) # Our custom tuning method.
Model Evaluation [1]
> glmnet
800 samples
1000 predictors
No pre-processing
Resampling: Cross-Validated (2 fold, repeated 5 times)
Summary of sample sizes: 400, 400, 400, 400, 400, 400, ...
Resampling results across tuning parameters:
alpha RMSE Rsquared MAE
0.000100 61.23760 0.002033470 52.85568
0.250075 61.83544 0.002543141 52.85747
0.500050 60.02843 0.002118089 52.03982
0.750025 59.16487 0.001651774 51.61048
1.000000 58.81907 0.002781994 51.40225
Tuning parameter 'lambda' was held constant at a value of 5
MAE was used to select the optimal model using the smallest value.
The final values used for the model were alpha = 1 and lambda = 5.
Model Evaluation [2]
- Now, let's predict on our testing data:
# Predict on testing data.
glmnet_prediction <- predict(glmnet_model, x_test)
# Check prediction accuracy.
postResample(pred = glmnet_prediction, # Our predicted values.
obs = y_test$V1) # Our expected vaues.
- This gives us a nice summary of the prediction:
RMSE Rsquared MAE
5.649786e+01 6.306793e-06 4.883650e+01
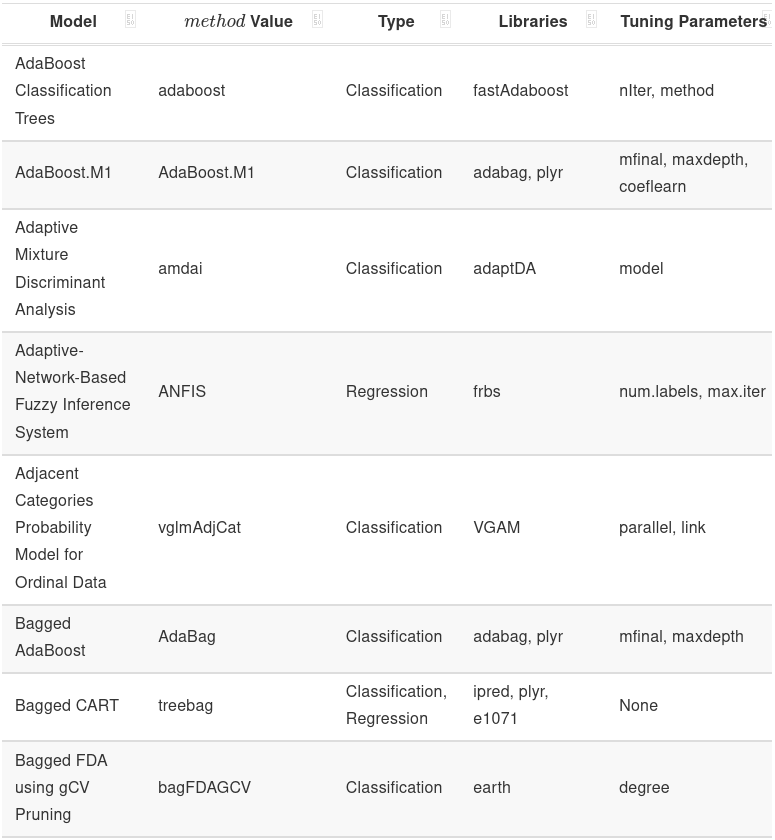
More Models
caretsupports a lot of models (last time I checked,238).- More examples are given in the file for you to try:
earthkerasxgboost
- You can also define your own models as well.
References
-
bigstatsRPackage:[1] https://github.com/privefl/bigstatsr [2] https://privefl.github.io/bigstatsr/articles/read-FBM-from-file.html
-
caretPackage:[1] https://topepo.github.io/caret/ [2] https://topepo.github.io/caret/available-models.html